2dSS is a web-server for visualising and comparing secondary structure predictions. It provides two main functionalities: 2D-alignment and compare predictions. The view 2D-alignment has been designed to visualise conserved secondary structure elements in a multiple sequence alignment (MSA). From this one can study the secondary structure content of homologous proteins (a protein family) and highlight its structural patterns. The compare predictions has been designed to compare the output of several secondary structure prediction tools, and check their accuracy when compared with real secondary structure elements extracted from 3D-structure. 2dSS provides a comprehensive representation of protein secondary structure elements, and it can be used to visualise and compare secondary structures of any prediction tool.
Welcome to the 2dSS web site
View 2D-alignment
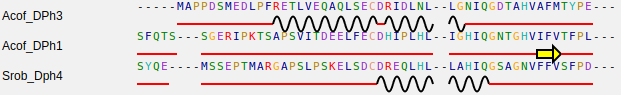
Given a multiple sequence alignment, representing a protein family, and the predicted SSEs of its constituent sequences, one can map each secondary structure elements onto a MSA position to obtain a 2D-alignment. The 2D-alignment highlights the secondary structure conservation in a set of related sequences, where is possible to see how secondary structure elements of homologous proteins are aligned.
Compare Predictions
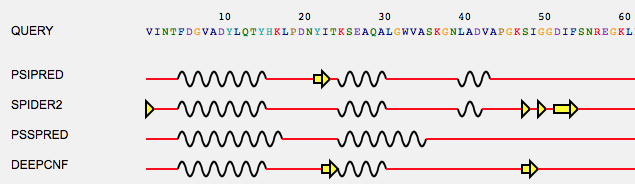
Compare predictions allows to compare secondary structure predictions of different tools, that is, you can identify differences in secondary structure element assignments among a set of prediction tools, and eventually compare those assignments to the real secondary structure elements extracted from the 3D structure.